
LIOU YIH-CHERNG
Associate Professor
Contact Information:
Department of Biological Sciences
National University of Singapore
14 Science Drive 4
Singapore 117543
6516 7711
6779 2486
dbslyc@nus.edu.sg
Academic Qualifications
D. Phil. (Queen’s, Canada), B. Sc. (NTOU, Taiwan)
Major Research Interests
The phosphorylation of specific proteins on Ser/Thr residues preceding proline is thought to be a major cellular signaling mechanism; however, very little is known about how the phosphorylation actually regulates protein function. Pin1 is an 18 kDa protein with two domains: the N-terminal WW domain (named after two invariant Trp residues, amino acids 1-39) and the C-terminal PPIase domain (amino acids 45-163). The WW domain acts as a binding module to bind its substrate via the phosphoserine (p-Ser) or phosphothreonine (p-Thr)-proline motifs in its substrates. The C-terminal PPIase domain is the catalytic domain to isomerize the conformational changes of specific pSer/Thr-Pro motifs. Unlike other peptidyl-prolyl isomerases (PPIases), Pin1 is a unique peptidyl isomerase that recognizes only the phosphorylated Ser/Thr motif preceding a proline residue. In addition, Pin1 is very prominent in isomerizing the cis-trans conformation of prolyl-peptidyl bonds in its substrates, resulting in regulation of their biological functions, suggesting the conformational changes mediated by Pin1 or Pin1-like isomerase may be crucial for the normal functioning of cells.
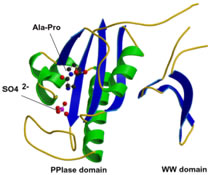
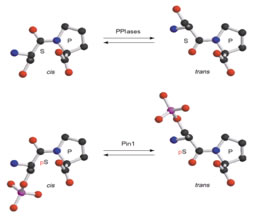
Our research directions lie in the area of how protein structure impacts signaling in the cells. The objectives are to characterize further the biology function of Pin1 and its downstream substrates. We are interested in understanding how the prolyl isomerase regulates its targets by changing the conformation on the pSer/Thr-Pro motifs in different organisms and in human diseases.
Selected publications
-
Li C, Xue C, Yang Q, Low BC & Liou YC. (2016) NuSAP governs chromosome oscillation by facilitating the Kif22 generated polar ejection force. Nat Commun. 7:10597 doi: 10.1038/ncomms10597.
-
Xi W, Gong X, Yang Q, Yu H, Liou YC. (2016) Pin1At regulates PIN1 polar localization and root gravitropism. Nat Commun. 7:10430. doi: 10.1038/ncomms10430.
-
Li C, Zhang Y, Yang Q, Ye F, Sun SY, Chen ES, Liou YC. (2016) NuSAP modulates the dynamics of kinetochore microtubules by attenuating MCAK depolymerisation activity. Sci Rep. 6:18773. doi: 10.1038/srep18773.
-
Ye GD, Sun GB, Jiao P, Chen C, Liu QF, Huang XL, Zhang R, Cai WY, Li SN, Wu JF, Liu YJ, , Xie YY, Chan EC, Liou YC, Li BA. (2016). OVOL2, an Inhibitor of WNT Signaling, Reduces Invasive ActivitiWu RSes of Human and Mouse Cancer Cells and Is Down-regulated in Human Colorectal Tumors. Gastroenterology. 2016 Mar;150(3):659-671.e16. doi: 10.1053
-
Xiao L, Xian H, Lee KY, Xiao B, Wang H, Yu F, Shen HM, Liou YC.(2015) Death-associated Protein 3 Regulates Mitochondrial-encoded Protein Synthesis and Mitochondrial Dynamics. J Biol Chem. 290(41):24961-74. doi:10.1074/jbc.M115.673343.
-
Gonzales KA, Liang H, Lim YS, Chan YS, Yeo JC, Tan CP, Gao B, Le B, Tan ZY, Low KY, Liou YC, Bard F, Ng HH. (2015). Deterministic Restriction on Pluripotent State Dissolution by Cell-Cycle Pathways. Cell. 162(3):564-79
-
Wong JJ, Li S, Lim EK, Wang Y, Wang C, Zhang H, Kirilly D, Wu C, Liou YC, Wang H, Yu F. (2013) A Cullin1-based SCF E3 ubiquitin ligase targets the InR/PI3K/TOR pathway to regulate neuronal pruning. PLoS Biol. 11(9). e1001657.
-
Chen HZ, Li L, Wang WJ, Du XD, Wen Q, He JP, Zhao BX, Li GD, Zhou W, Xia Y, Yang QY, Hew CL, Liou YC*, Wu Q* (2012) Prolyl isomerase Pin1 stabilizes and activates orphan nuclear receptor TR3 to promote mitogenesis. Oncogene. 31(23):2876-87 (*Co-corresponding author)
-
Ye F, Tan L, Yang Q, Xia Y, Deng LW, Murata-Hori M, Liou YC (2011) HURP Regulates Chromosome Congression by Modulating Kinesin Kif18A Function. Curr Biol. 21:1584-91.
-
Liou YC*, Zhou XZ*, Lu KP* (2011). Prolyl isomerase Pin1 as a molecular switch to determine the fate of phosphoproteins. Trends Biochem Sci. 36:501-14 (*Co-corresponding authors)
